After the inception of next-generation DNA sequencing, RNA sequencing (RNA-Seq) was developed, which provided a great tool for transcriptome profiling. However, conventional RNA-Seq still has much room for improvement. For example, conventional RNA-Seq is hampered by inaccuracy from exponential PCR amplification and sequence-dependent bias during sample preparation and sequencing.
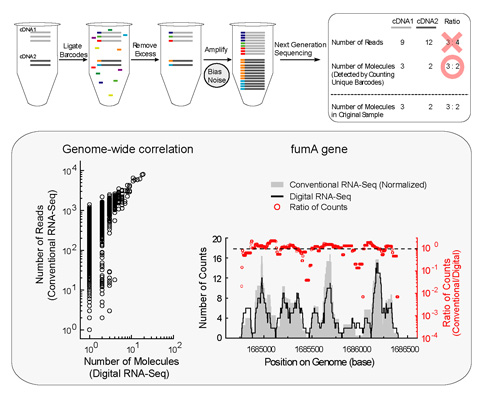
We have developed a simple strategy for mitigating these complications, allowing truly digital RNA-Seq. Following reverse transcription, a large set of barcode sequences is added in excess, and nearly every cDNA molecule is uniquely labeled by random attachment of barcode sequences. After PCR, we applied deep sequencing to read the barcode and cDNA sequences. Rather than counting the number of reads, RNA abundance is measured based on the number of unique barcode sequences observed for a given cDNA sequence. We optimized the barcodes to be unambiguously identifiable, even in the presence of multiple sequencing errors. This method allows counting with single-copy resolution despite sequence-dependent bias and PCR-amplification noise, and is analogous to digital PCR but amendable to quantifying a whole transcriptome. We demonstrated transcriptome profiling of Escherichia coli with more accurate and reproducible quantification than conventional RNA-Seq. This technique is very useful for single cell analysis which requires high accuracy and sensitivity.
References:
Shiroguchi, Katsuyuki; Jia, Tony Z.; Sims, Peter A.; Xie, X. Sunney. "Digital RNA Sequencing Minimizes Sequence-dependent Bias and Amplification Noise with Optimized Single-molecule Barcodes," PNAS 109:1347-1352 (2012).
|

