Like enzymology in general, single-molecule enzymology has been limited and facilitated by assay developments. Recent advances in single-molecule enzymatic assays have profoundly changed how biochemical reactions are studied, revealing new insights into established problems and opening new lines of investigation. To that end, our group has developed single molecule fluorescence and flow stretching enzymatic assays.
Single Molecule Fluorescence Assays by Fluorescent Measurements
Here we list the single-molecule enzymatic turnover assays by fluorescence detection:
-
Assays with fluorescent active sites [1], as demonstrated for cholesterol oxidase in the left figure above.
-
Assays with fluorescently labeled substrates, such as ATP or dNTP, which require either low substrate concentrations [2] or zero-mode waveguide to suppress the strong background fluorescence signal [3]. This latter method has been used for single molecule DNA sequencing.
-
Assays with FRET pairs that report conformational changes triggered by enzymes, such as staphylococcal nuclease [4], a ribozyme [5], T4 Lysozyme [6].
-
Assays based on rotational [7] and translational [8] trajectories of molecular motors by recording movies of fluorescent labels.
-
Assays based on intensity changes of fluorophores that are associated with conformational changes, such as fluorescence quenching in a DNA polymerase upon substrate dNTP binding [9].
-
Assays based on a fluorogenic substrate converted to a fluorescent product molecule by an enzymatic reaction (see right figure above), which was first proposed and attempted for horseradish peroxidase [10]. Because fluorophores are continuously replenished and diffuse away from the excitation volume, this assay circumvents the photobleaching problem that hampers all the other assays, and provides extremely long time traces [11,12].
Flow Stretching Assay
We have exploited an extremely simple assay based on flow-stretched DNA to monitor the
activity of many individual nucleic acid enzymes on DNA simultaneously [13-15]. Individual DNA
molecules are attached at one end of the glass surface of a flow cell. When a laminar flow is applied
above the surface, the DNA molecules are stretched. If beads are attached to the free ends
of the DNA, the molecules are stretched to a uniform tension by the drag force on the beads,
allowing careful measurement of the DNA lengths by tracking the positions of the beads with
an inexpensive CCD camera (accuracy ~10 nm for seconds, ~100 nm for minutes).
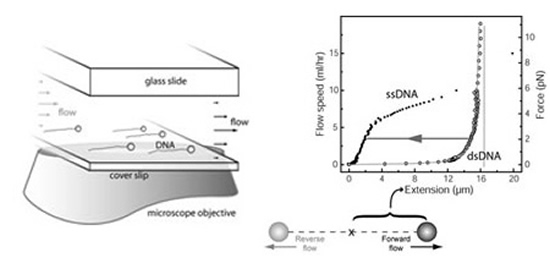
The force-extension curves of a single-stranded (ssDNA) and a double-stranded (dsDNA) l phage
DNA (48,502 nt) are shown. Much of the pioneering work of studying the mechanical properties of
individual DNA molecules is done in Carlos Bustamante's Lab at UC Berkeley [164]. The coiling of ssDNA causes it to be shorter than dsDNA at low stretching forces (<6 pN). Consequently, a (enzymatic)
conversion between dsDNA and ssDNA can be monitored by the variation of the DNA length at
a constant stretching force [17,18].
For example, we used this multiplexed single-molecule method to study the enzymatic activity of λ
Exonuclease [13], a homotrimeric, ringshaped enzyme required for recombination in bacteriophage λ,
which degrades each strand of duplex DNA in the 5' to 3' direction.
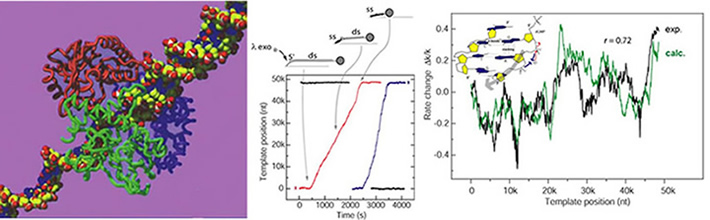
Some of the individual λ exonuclease enzyme molecules digest processively all 48knt of the λ phage
DNA substrate. The digestion rate of the individual enzymes exhibits large fluctuations. These
fluctuations are partially dependent on the sequence of the λ DNA. We calculate the variations
in the enzymatic rate caused by the modulation on the free energy required to melt the cleaved
bases (green trace*) and compare it to the average of multiple single-molecule rate traces (black
trace). The high degree of correlation identifies the base melting as a rate-limiting step in the
catalytic cycle.
We note that many single molecule assays are based on optical tweezers and magnetic
tweezers on nucleic acid enzymes and molecule motors, which were pioneered by the David Bensimon/Vincent Croquette [18], Steve Block [19], Carlos Bustamante [17], and Toshio Yanagida [2] groups.
Single-Molecule Transcription Assay
We have developed a high-throughput in vitro single-molecule assay to monitor repetitive stochastic transcription events in real time on individual DNA templates [20]. We use a nucleic acid stain, SYTO RNASelect, that is nonfluorescent at 530 nm but becomes fluorescent upon binding to RNA. It has been used to detect RNA in the presence of DNA [21]. An Argon laser line at 488 nm is used to excite SYTO RNASelect in a total internal reflection fluorescence (TIRF) microscope. We collect fluorescence at 530 nm and record time-lapse movies with a CCD camera. In the presence of the dye, a single nascent mRNA becomes visible, and its fluorescence intensity increases with the mRNA length.
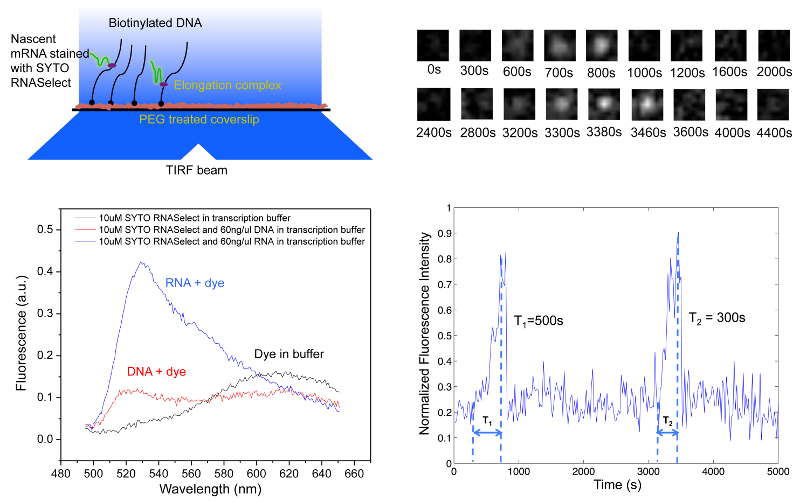
We tether DNA templates containing a promoter on the passivated surface of the flow cell through biotin-streptavidin linkage. After we infuse a mixture of RNA polymerase, NTPs and SYTO RNASelect into the flow cell, the fluorescence intensity of many spots in the field of view linearly ramps up due to transcription elongation, which is followed by abrupt disappearance upon transcription termination. "Blinking" of fluorescence occurs when multiple transcripts are produced from a template. By recording fluorescent movies, we could monitor transcription elongation in real time as nascent mRNAs are produced on hundreds of templates simultaneously.
Transcription is known to generate DNA positive supercoiling ahead of the elongation complex and negative supercoiling behind it when the free spiral of the bulky complex around the DNA is hindered by the frictional drag on the complex [22,23]. This single-molecule assay allows us to investigate the effects of supercoiling on transcription initiation and elongation in a clean and controlled system.
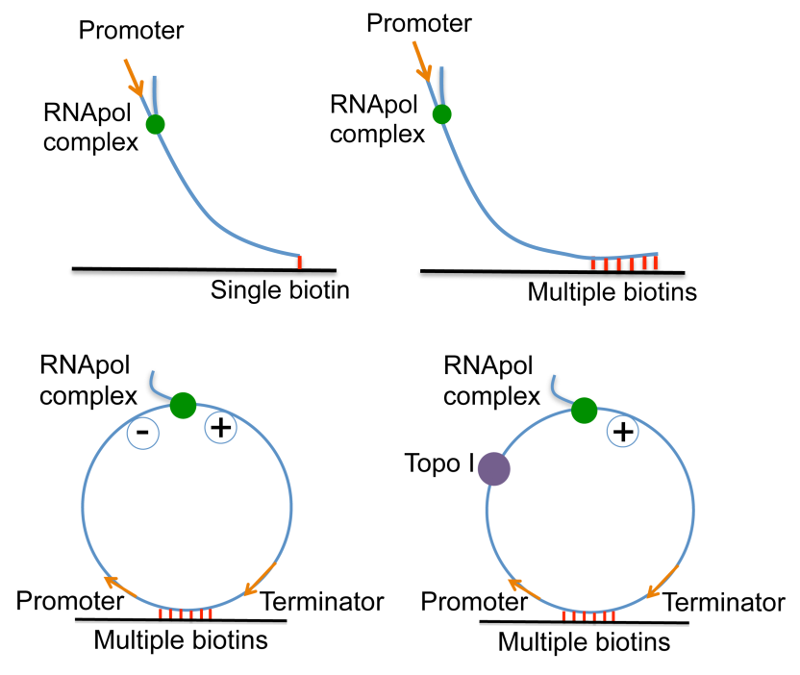
In order to examine the effect of positive supercoiling buildup on transcription elongation, we design 12 kb-long linear DNA templates with a promoter on the 5' end, and single or multiple biotinylated nucleotides on the 3' end. On the single-biotin template, supercoiling cannot accumulate since DNA can rotate around its single linkage to the surface. In contrast, on the multiple-biotin template, DNA cannot rotate around its multiple linkages, thus positive supercoiling would accumulate downstream of the elongation complex. We have shown that positive supercoiling generated by transcription slows down elongation [20].
In order to examine the effect of positive supercoiling on transcription initiation, we design a circular template and tether it to the surface with multiple biotin-streptavidin linkages. The circular template consists of a promoter, a 12 kb-long transcribing sequence and a terminator. In the presence of E. coli Topoisomerase I, negative supercoiling generated by transcription is continuously removed, and positive supercoiling accumulates on the circular template as multiple transcripts are made from the template. We have shown that positive supercoiling generated by transcription stops initiation [20].
We note our assay is different from other existing in vitro transcription assays using single-molecule manipulation or single-molecule fluorescence imaging. Our assay uses RNA staining so that the elongation process on templates with any sequence can be easily monitored for multiple rounds of transcription on each template. Meanwhile, this is a high-throughput measurement because hundreds of templates in one field of view can be monitored simultaneously. This assay will be generally useful for studying other questions in transcription, such as pausing and termination kinetics.
References
[1] H.P. Lu, L. Xun, X.S. Xie. Science 282, 1877 (1998).
[2] Funatsu T., et al. Nature 374, 555 (1995).
[3] M.J. Levene, J. Korlach, S.W. Turner, M. Foquet, H.G. Graighead, W.W. Webb. Science 299, 682–686 (2003).
[4] T. Ha, et al. PNAS 96, 893–898 (1999).
[5] X. Zhuang, et al. Science 288, 2048–2051 (2000).
[6] C. Yu, D. Hu, E.R. Vorpagel, H.P. Lu. J. Phys. Chem. B. 107, 7947 (2003).
[7] R. Yasuda, H. Noji, K. Kinosita, M.Yoshida. Cell, 93, 1117 (1998).
[8] A. Yildiz, J. N. Forkey, S. A. McKinney, T. Ha, Y. E. Goldman, P.R. Selvin. Science 300, 2016, (2003).
[9] G. Luo, et al., PNAS 104, 12610–12615 (2007).
[10] L. Edman et al. Chem. Phys. 247, 11–22 (1999).
[11] B. English, et al. Nat. Chem. Biol. 2, 87–94 (2006).
[12] K. Velonia, O. Flomenbom, D. Loos, S. Masuo, M. Cotlet, Y. Engelborghs, J. Hofkens, A.E. Rowan, J. Klafter, R.J.M. Nolte, F.C. de Schryver. Angew. Chem. Int. Ed. (2004) 43, 2–6 (2005).
[13] A.M. van Oijen, P.C. Blainey, D.J. Crampton, C.C. Richardson, T. Ellenberger, X.S. Xie. Science 301, 1235 (2003).
[14] C.M. Schroeder, P.C. Blainey, S. Kim, X.S. Xie. In "Single Molecule Techniques A Laboratory Manual" edited by P.R. Selvin and T. Ha, p 461-492, Cold Spring Harbor Laboratory Press, 2008.
[15] S. Kim, P.C. Blainey, C.M. Schroeder, X.S. Xie. Nature Methods, 4, 397-399 (2007).
[16] S.B. Smith, L. Finzi, C. Bustamante. Science 258, 1122 (1992).
[17] G.J.L. Wuite, S.B. Smith, M. Young, D. Keller, C. Bustamante. Nature 404, 103 (2000).
[18] B. Maier, D. Bensimon, V. Croquette. PNAS 97, 12002 (2000).
[19] K. Svoboda, C.F. Schmidt, B.J. Schnapp, S.M. Block. Nature, 365, 721 (1993).
[20] S. Chong, C. Chen, H. Ge, X.S. Xie. Cell 158, 314-326 (2014).
[21] C. Kannemeier et al. PNAS 104, 6388-6393 (2007).
[22] L.F. Liu, J.C. Wang. PNAS 84, 7024-7027 (1987).
[23] Y.P. Tsao, H.Y. Wu, L.F. Liu. Cell 56, 111-118 (1989).
