High-throughput sequencing is rapidly changing the ways that biological
questions are addressed. There is a growing need for sequencing a single
cell, as genetic variations unique to individual cells are often lost in a
population average. So far there has not been a single molecule approach
that can read the entire genome of a single cell. Instead, one has to
rely on whole genome amplification. However, this has been hindered by
strong amplification bias, resulting in a large fraction of the genome
being undetectable.
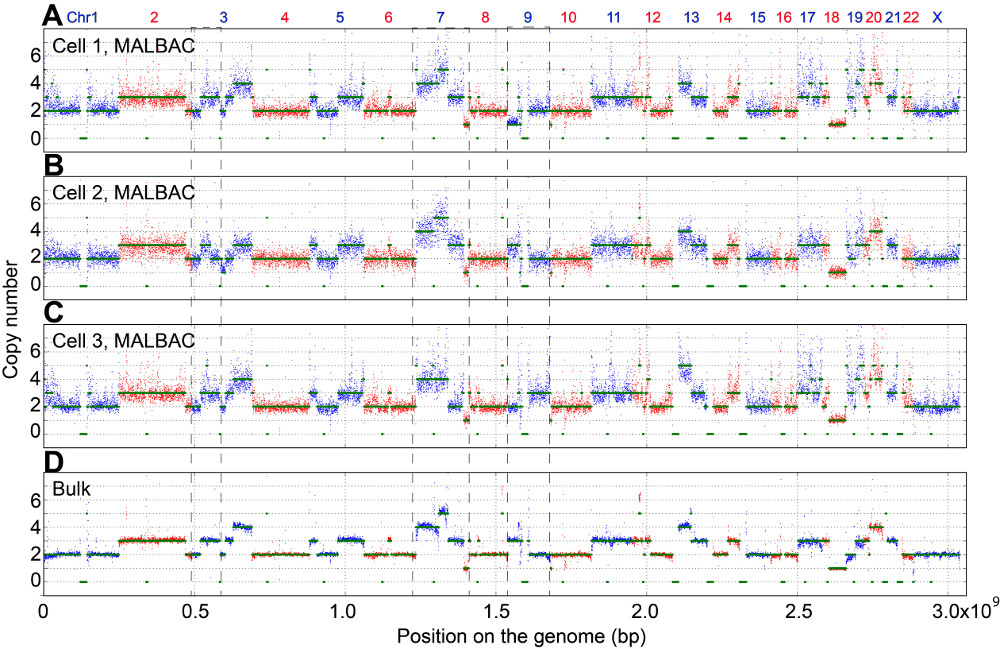
Fig. 1. Single cell copy number variations (CNVs)
We have recently developed a technique for whole genome amplification
that enables sequencing the genome of a single cell with high coverage.
The technique—Multiple Annealing and Looping-Based Amplification Cycles
(MALBAC)—uses a uniform, linear pre-amplification of DNA prior to
exponential amplification by PCR. The unprecedented uniformity provided
by MALBAC allows accurate detection of both single point mutations and
copy number variations of individual cells. We used this ability to
study the generation of new-born mutations in a cancer cell line and
were able to measure the mutation rate directly.
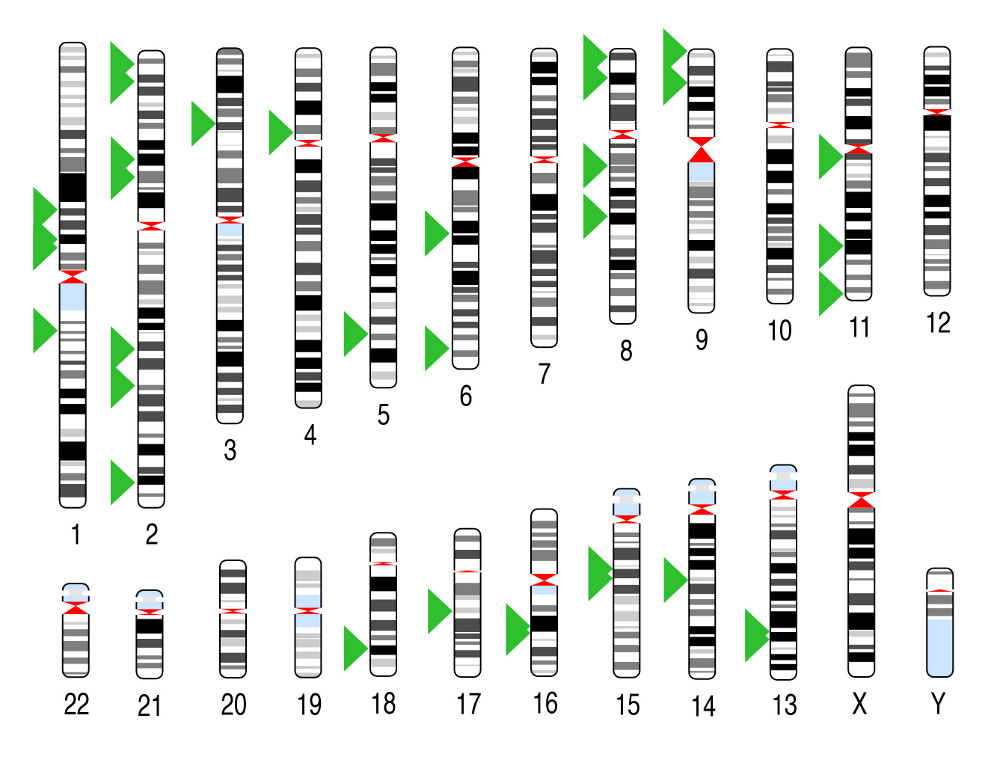
Fig. 2. A single cancer cell's new-born single nucleotide variations (SNVs) that do not exist in the bulk. No false positives are detected
As one of the first applications, we applied MALBAC to sequence ~100
sperm cells of an individual, in collaboration with colleagues of BIOPIC at Peking University.
During meiosis, crossover events occur,
creating genetic diversity by mixing paternal and maternal DNA. By
observing SNPs in a single sperm, we can identify the position of
crossover events and occasional segregation errors of chromosomes. We are
able to obtain the spatial distribution of recombination events along the
chromosomes with the highest resolution. Such an approach can be useful
in probing the origin of infertility.
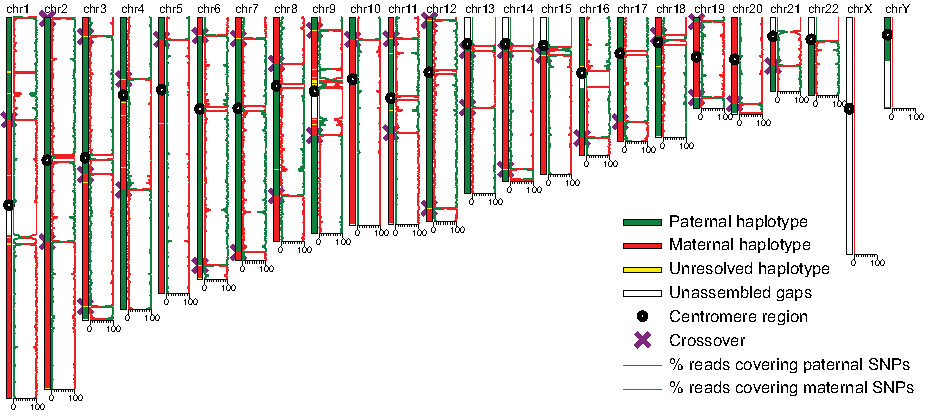
Fig. 3. Recombination events in a single sperm cell
With our collaborators, we are applying MALBAC to cancer diagnostics with circulating tumor cells,
as well as to early prenatal diagnostics with fetal cells in the mother's
blood. In both cases only a few cells are available
for analysis.
References:
Zong, Chenghang; Lu, Sijia; Chapman, Alec R.; Xie, X. Sunney. "Genome-Wide Detection of Single-Nucleotide and Copy-Number Variations of a Single Human Cell," Science 338:1622-1626 (2012).
Lu, Sijia; Zong, Chenghang; Fan, Wei; Yang, Mingyu; Li, Jinsen; Chapman, Alec R.; Zhu, Ping; Hu, Xuesong; Xu, Liya; Bai, Fan; Qiao, Jie; Tang, Fuchou; Li, Ruiqiang; Xie, X. Sunney. "Probing Meiotic Recombination and Aneuploidy of Single Sperm Cells by Whole-Genome Sequencing," Science 338:1627-1630 (2012).
|

