|
|
|
| Mechanism of Transcriptional Bursting in Bacteria |
Transcription of highly expressed genes has been shown to occur in stochastic bursts in bacteria (1-4). But the origin of such ubiquitous phenomenon was not understood previously. Through a series of single-molecule in vitro and live-cell experiments, we have uncovered the mechanism of this ubiquitous phenomenon.
It has been previously shown that E. coli nucleoid-associated proteins serve as anchoring points and create ~400 topologically constrained loops on the chromosomal DNA (5-7). Transcription in a loop generates positive supercoiling ahead of the RNA polymerase, and negative supercoiling behind the polymerase. While negative supercoiling formed during transcription is immediately removed by topoisomerase I, the activity of gyrase which is responsible for removing positive supercoiling, is not as efficient. Since on average there is one gyrase molecule per chromosomal DNA loop in living E. coli cells, the stochastic effect of gyrase-DNA interaction is prominent. We hypothesize that a gene stochastically switches between on and off states due to release and accumulation of positive supercoiling in a chromosomal DNA loop upon gyrase binding and dissociation, respectively (Fig. 1).
|
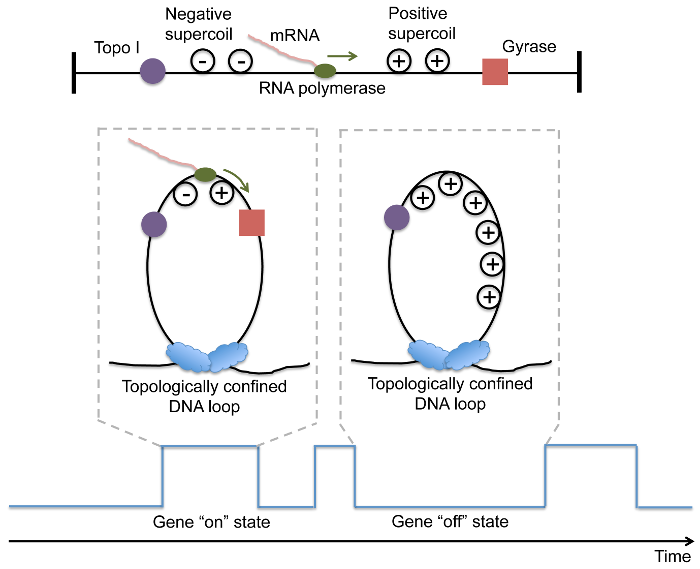
Figure 1: DNA Topology vs. Transcriptional Bursting in E. coli
Transcription of DNA into RNA by RNA polymerase results in positive supercoiling in front of the polymerase and negative supercoiling behind the polymerase. Topoisomerase I relieves negative supercoiling; gyrase relieves positive supercoiling (higher schematic). In the absence of a positive supercoiling stall force, RNA synthesis can proceed (lower-left). As positive supercoiling accumulates or in the absence of DNA gyrase, transcription is inhibited (lower-right). This alternation between active and inactive transcription accounts for the "on" and "off" behavior that characterizes transcriptional bursting (lowest schematic).
|
|
In order to test this hypothesis in a clean and controlled fashion, we developed a high-throughput in vitro single-molecule assay to follow transcription in real time on individual DNA templates with controlled supercoiling levels (8). This is a fluorescence-imaging based assay that uses RNA staining, which allows measurement of transcription elongation and initiation rates. Using this assay, we showed that positive supercoiling buildup on the DNA by transcription slows down transcription elongation and eventually stops transcription initiation. Transcription can be resumed upon gyrase binding to the DNA segment. We further determined the Kd and the rate constant of gyrase-DNA binding in vitro, and found that are consistent with the "off" and "on" periods of transcriptional bursting observed in live E. coli cells (1).
We measured the mRNA copy number distribution in a population of isogenic E. coli cells using single-molecule mRNA fluorescence in situ hybridization (FISH) (3), and found that the distribution can be fitted well by the "Poisson with zero spike" distribution. The fitting allows determination of the on/off duty cycle ratio of transcriptional bursting. We examined the fully induced lac operon and another 18 highly transcribed E. coli genes, and found all of them show a decreased on/off duty cycle upon gyrase inhibition (lowering of intracellular gyrase concentration), as predicted by the two-state model (8). This result demonstrates the ubiquity of the local supercoiling and gyrase activity effect on transcriptional bursting.
Together, these findings provide definitive proof for our hypothesis. In summary, through a series of single-molecule in vitro and live-cell experiments, we proved that transcriptional bursting of highly expressed genes in bacteria is primarily caused by gyrase dissociation from and reversible binding to the chromosomal DNA segment, changing the supercoiling level of the DNA segment. This work draws a direct line between DNA mechanics and stochastic gene expression.
|
|
References
- Golding, I., Paulsson, J., Zawilski, S.M., and Cox, E.C. (2005). Real-time kinetics of gene activity in individual bacteria. Cell 123, 1025-1036.
- So, L.H., Ghosh, A., Zong, C., Sepulveda, L.A., Segev, R., and Golding, I. (2011). General properties of transcriptional time series in Escherichia coli. Nat Genet 43, 554-560.
- Taniguchi, Y., Choi, P.J., Li, G.W., Chen, H., Babu, M., Hearn, J., Emili, A., and Xie, X.S. (2010). Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329, 533-538.
- Zong, C., So, L.H., Sepulveda, L.A., Skinner, S.O., and Golding, I. (2010). Lysogen stability is determined by the frequency of activity bursts from the fate-determining gene. Mol Syst Biol 6, 440.
- Hardy, C.D., and Cozzarelli, N.R. (2005). A genetic selection for supercoiling mutants of Escherichia coli reveals proteins implicated in chromosome structure. Mol Microbiol 57, 1636-1652.
- Postow, L., Hardy, C.D., Arsuaga, J., and Cozzarelli, N.R. (2004). Topological domain structure of the Escherichia coli chromosome. Gene Dev 18, 1766-1779.
- Wang, W., Li, G.W., Chen, C., Xie, X.S., and Zhuang, X. (2011). Chromosome organization by a nucleoid-associated protein in live bacteria. Science 333, 1445-1449.
- Chong, S., Chen, C., Ge, H., and Xie, X.S. (2014). Mechanism of Transcriptional Bursting in Bacteria. Cell 158, 314-326.
|
|
|
 |
|
