Painting a specific chromosome with CRISPR/Cas9 for live-cell imaging
Visualization of chromosome shapes and dynamics in a live cell is highly desirable and necessary in many areas of cell biology. During interphase, each chromosome exists in its own territory in the nucleus, which can be imaged by fluorescence in situ hybridization (FISH) using sequence-specific probes of different colors. However, such chromosome painting has only been possible in fixed cells, and is not suitable for dynamic monitoring of live cells. Recent development of clustered regularly interspaced short palindromic repeats/CRISPR-associated proteins (CRISPR/Cas) has provided a powerful tool for live cell imaging of genomic loci. However, the labeling achieved by this method is usually restricted to the genomic loci that consist of repetitive sequences, and has not been attempted to track an entire chromosome in a live cell. In collaboration with Wensheng Wei's group at BIOPIC, we designed a new strategy using a large number of sgRNAs targeting mainly the non-repetitive regions of the chromosome. The specific labeling makes it possible to paint an entire chromosome in a live cell.
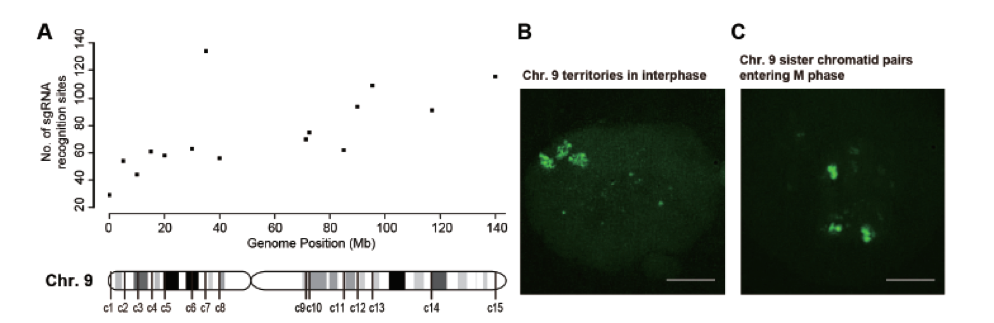
Figure 1. (A) Scatter plot for numbers of sgRNA-binding sites in each cluster of 5 Kb width across human chromosome 9. (B) Maximum intensity projection of EGFP images recorded by N-SIM at the S phase of the cell cycle. (C) Maximum intensity projection of EGFP images recorded by N-SIM in the prophase of mitosis.
Figure 2 shows chromosome dynamics at a single chromosome level for a cell in the S phase. From the late S phase to M phase, in addition to the three chromosome 9 spots, the nucleoli exhibit unintended EGFP signal, likely due to nonspecific binding of dCas9-EGFP protein with small RNAs in the nucleoli. During the data collection period, EGFP signal from the nucleoli faded away due to the disappearance of the nucleoli in the M phase, while chromosome 9 fluorescent signal became stronger due to chromosome condensation. After cell division, the two daughter cells maintained strong fluorescence signals from the replicated chromosomes, indicating that chromosome labeling is kept in the daughter cells.
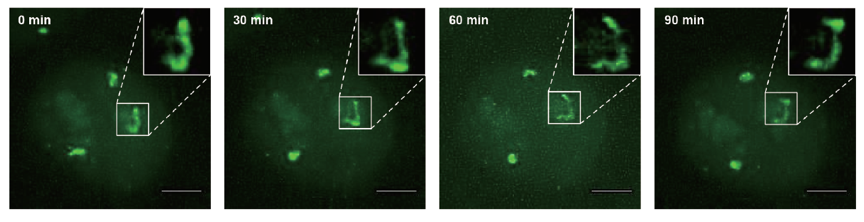
Figure 2. Dynamics of the three copies of chromosome 9 in S phase in a live HeLa cell.
By introducing hundreds of specific and non-repetitive sgRNAs in a human cell, we are able to paint an entire chromosome in a live cell for fluorescent imaging. We have visualized the spatial arrangements of homologous chromosomes and sister chromatids and tracked the movement of a particular chromosome in dividing cells. Our method will facilitate studies of functional organization of chromosomes, interactions among different chromosome regions, and long-term chromosomal dynamics in live mammalian cells.
References:
| 1. |
Zhou, Yuexin, Ping Wang, Feng Tian, Ge Gao, Lei Huang, Wensheng Wei, and X. Sunney Xie. "Painting a specific chromosome with CRISPR/Cas9 for live-cell imaging." Cell Research (2017). |
