|
|
|
| Allostery Through DNA |
DNA encodes genetic information as a string of base sequences in a double-helical structure. The base sequence encodes not only genes whose expression determine cell functions, but also provides specific binding sites for proteins, called transcription factors, whose binding and unbinding to DNA control gene expression levels based on environmental signals.
We have uncovered a physical property of DNA, called allostery, which affects DNA-protein interactions. Allostery means that binding of a molecule on one part of a macromolecule affects the binding of another molecule at a distal site of the macromolecule. Such a long-range effect within a macromolecule has been extensively studied for enzymes or proteins (1,2), but has not been quantitatively characterized or mechanically (3-7). If DNA is completely rigid or completely floppy, the binding of one protein would be independent of the presence of another DNA-bound protein nearby.
We measured how long a protein binds on its DNA-binding site before it falls off, or the mean residence time, which is also the inverse of the dissociation rate constant (koff). The protein was labeled with a fluorescent dye. By using single-molecule fluorescence microscopy based on total internal reflection illumination, we could monitor individual protein dissociation events from a few hundred DNA templates simultaneously. We designed DNA sequences such that there are two protein-binding sites with varying separation (L). One binding site was for the fluorescently labeled protein and the other binding site was for a different type of DNA-binding protein that is not labeled (Fig. 1A for a fluorescently labeled DNA-binding domain of glucocorticoid receptor, or GRDBD on its binding site, GRE, and BamHI).
|
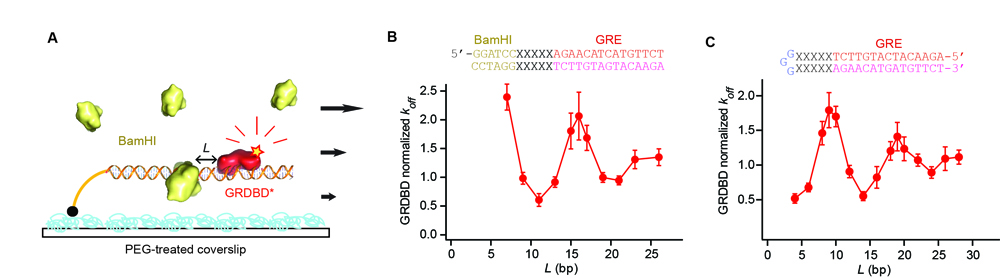
Figure 1
|
|
Our experiment showed that two DNA-binding proteins can affect each other's binding affinity to DNA at a distance in such a way that their binding is either simultaneously enhanced or reduced (Fig. 1B). The long-range effect not only varies as a function of the distance between two protein-binding sequences, but also oscillates between the enhancement and reduction with the magic 10 base period, dictated by the famous double helical structure of DNA. It is a rather large effect (about five-fold change) that decays within ~15 base pairs, previously hidden in conventional ensemble experiments. Also, such an allosteric property of DNA is general, true for any protein pairs we examined (8). This is significant as multiple proteins, such as transcription factors and RNA polymerase, often bind close to each other on genomic DNA to carry out their cellular functions in concert.
We performed various control experiments to verify that such a change truly comes from DNA allostery. The key experiment was performed with DNA hairpin: we designed DNA to have a hairpin loop (a single-stranded, unpaired region of DNA) at one end of the double-stranded DNA and placed a binding site for the fluorescently labeled protein near the hairpin loop. We discovered that the residence time of the protein was also altered by its distance to the hairpin loop (Fig. 1C). We know that a hairpin loop creates distortion in the double helical structure of DNA. This experiment ruled out the possibility of direct protein-protein interaction as the origin of the observed behavior and suggests that the distortion of DNA double helix can propagate along the DNA and affect binding of a protein at a distal site.
|

Figure 2
|
|
To understand the mechanism for the long-range effect through DNA, think of the DNA-bound proteins as monkeys and DNA as a monkey bar (Fig. 2). Two monkeys hang on a monkey bar, trying hard to hold their grip. Interestingly, the time a monkey stays on the bar is affected by the presence of another monkey hanging nearby. Imagine the binding of the first monkey widens the separation between the blue and red strands (analogy to the major groove width). Such distortion of the rope structure will propagate along the rope. If the second monkey sits nearby at a place where the gap between the blue and red strand is widened, it is easier to stay put as it doesn't have to work as hard to widen the two strands! On the other hand, if the second monkey sits at a place where the gap between the two strands is narrowed, it is difficult to stay put there!
We demonstrated that DNA allostery indeed affects gene expression inside live cells and transcription factor binding near nucleosomes. Hence, DNA allostery is physiologically important, and future studies must take this into account.
References
- J. Monod, J. Wyman, J. P. Changeux, J. Mol. Biol. 12, 88 (1965).
- D. E. Koshland, G. Némethy, D. Filmer, Biochemistry 5, 365 (1966).
- F. M. Pohl, T. M. Jovin, W. Baehr, J. J. Holbrook, Proc. Natl. Acad. Sci. U.S.A. 69, 3805 (1972).
- M. Hogan, N. Dattagupta, D. M. Crothers, Nature 278, 521 (1979).
- R. Moretti et al., ACS Chem. Biol. 3, 220 (2008).
- E. F. Koslover, A. J. Spakowitz, Phys. Rev. Lett. 102, 178102 (2009).
- Hernan G. Garcia et al., Cell Reports 2, 150 (2012).
- S. Kim et al., Science 339, 816 (2013).
|
|
|
 |
|
